LaBaer Lab | Home
FlexGene
Development Team: Michael Fiacco and Preston Hunter
Data management has been identified as a crucial issue in all large scale experimental projects. In this type of project, many different researchers manipulate multiple objects in different locations; thus, unless complete and accurate records are maintained, it is extremely difficult to understand exactly what has been done, when it was done, who did it, and what protocol was used.
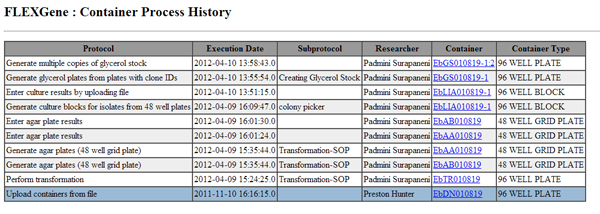 We have developed a laboratory information management system (LIMS) called FLEXGeneDB (FLEX) (publication) for tracking high throughput cloning processing, which is a major production pipeline in the lab. The FLEX system tracks every step in the production process and integrates with liquid handling robots to eliminate human errors. It maintains a detailed history for each plate and sample by capturing processing parameters, protocols and analytical results for the complete life cycle of the sample (see figure at right,screenshot of a plate history GUI) . FLEX is capable of managing clones produced outside our lab that require further processing such as transferring to different vectors or sequence validation.
We have developed a laboratory information management system (LIMS) called FLEXGeneDB (FLEX) (publication) for tracking high throughput cloning processing, which is a major production pipeline in the lab. The FLEX system tracks every step in the production process and integrates with liquid handling robots to eliminate human errors. It maintains a detailed history for each plate and sample by capturing processing parameters, protocols and analytical results for the complete life cycle of the sample (see figure at right,screenshot of a plate history GUI) . FLEX is capable of managing clones produced outside our lab that require further processing such as transferring to different vectors or sequence validation.
Processing of biological samples in FLEX is accomplished by establishing workflows that parallel the processing steps completed in the laboratory. Samples (plates) are grouped on a project/workflow basis, where a project is defined by the purpose of cloning or the organism (see figure at left for overview of conceptual structure and relations between most important FLEX components). FLEX has a GUI that allows users to get full access to the information tracked by the application, including but not limited to files produced by laboratory robots.
The majority of successful clones processed through FLEXGeneDB eventually enter the in-house sequence validation software ACE for sequence validation and then to the repository system called DNASU for storage and distribution. FLEXGeneDB transfers data between these three applications that support clone processing and distribution in the lab.

FLEXGeneDB is a three-tier web application that can be run on Linux or Windows XP servers. The application’s GUI has been implemented in JSP and javascript and tested for compatibility with common browsers. A relational database (Oracle) is used for back-end data storage. The application layer is implemented in java, and result files are stored on the server hard drive. Some time-consuming operations are handled asynchronously: the application runs the job in the background and informs the user by e-mail once the task has been finished. The email summarizes the job request as well as the associated results.
FLEXGeneDB was launched in 2002. The Center’s bioinformatics team continues to add new features over time. At present, FLEX database has tracked over two million samples and ~24,000 containers that have flowed through the lab.
Publications
Brizuela L, Richardson A, Marsischky G, Labaer J. The FLEXGene repository: exploiting the fruits of the genome projects by creating a needed resource to face the challenges of the post-genomic era. Arch Med Res. 2002 Jul-Aug;33(4):318-24. PMID: 12234520

