LaBaer Lab | Home
High Throughput Cloning Team

Team Leader: Andrea Throop, Ph.D.
Team:Kevin Peasley and Hannah Johnson
The High Throughput (HT) Cloning/Production Team within the Center for Personalized Diagnostics at the Biodesign institute is responsible for several projects and pipelines. These include the full length sequence validation of over 150,000 Protein Structure Initiative (PSI) clones from NIGMS-funded PSI centers (see pipeline to right). In addition, our team does all of the recombinational cloning and clone validation projects both within the center and with numerous collaborators. All plasmids validated and created by the cloning team can be found in the DNASU Plasmid Repository.
Within the center, each step of our cloning, sequencing and distribution process is tracked by laboratory information management systems (LIMS) and facilitated by robots. Our processing pipeline is shown below.Roll over each image to see a zoomed-in view of the robot or process results.
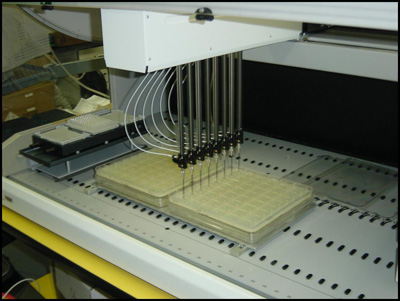
Transformation and Colony Picking
We receive plates of plasmids to be distributed through DNASU either as a 96-well plate of DNA or as glycerol stocks in phage resistant bacteria. When we receive DNA, we perform a transformation on the DNA in the entire 96-well plate and then plate the bacterial transformation on a patented 48-well agar plate using the FX liquid handling robot (see image). Individual colonies are picked into a deep well block using a K6 colony picker. Roll over the image to see the colonies growing on the 48-well plate after incubating overnight at 37 degrees Celsius.

Sanger Sequencing
After picking a colony and growing overnight in liquid culture, we copy the plate into a storage copy and a working copy. Using the FX liquid handling robot, we purify the DNA in high throughput for Sanger sequencing. Roll over the image to see a view of the chromatograph from the Sanger Sequencing machine.
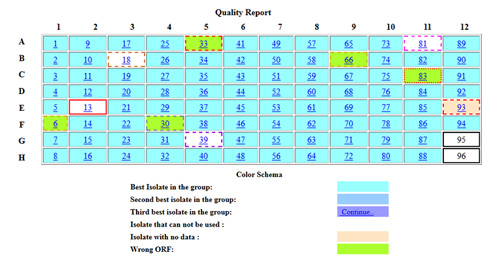
Sequence Evaluation using ACE
Once sequenced, we use our Automated Clone Evaluation (ACE) software for sequence analysis. ACE uses artificial intelligence (AI) to analyze the raw sequence data by assembling the actual sequence of an insert into a contig and comparing it to its expected sequence. Each difference between actual and expected sequences is recorded as a discrepancy object including its position, bases, and sequence confidence. Acceptance and rejection are determined by counting the number of each discrepancy type and its confidence and comparing that against what is considered acceptable for a particular project. The discrepancies listed for a particular plasmid typically come from our analysis using ACE. Roll over the ACE screen shot for an example alignment.
LIMS systems
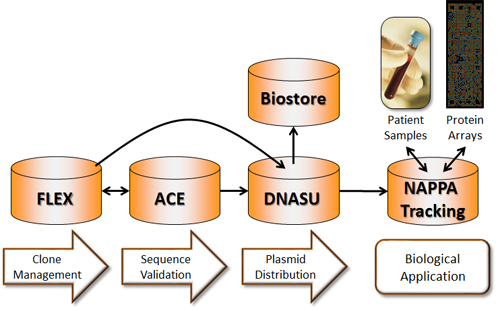 Efficient and accurate management of HT cloning relies on a solid informatics pipeline, which supports all aspects of DNA manipulation, sample handling, sequence validation, and protein and microarrays. Our facility tracks and analyzes each step of the cloning and sequencing project using Laboratory Information Management Systems (LIMS) developed in our laboratory. These LIMSs generate bar codes and text files that communicate directly with our robots and parallel every step of the Gateway recombinational cloning system from primer design through cloning, and ultimately sequence verification of full-length clones, thus ensuring a seamless flow of information between robots and LIMSs throughout (see figure). You can find more details about these systems here.
Efficient and accurate management of HT cloning relies on a solid informatics pipeline, which supports all aspects of DNA manipulation, sample handling, sequence validation, and protein and microarrays. Our facility tracks and analyzes each step of the cloning and sequencing project using Laboratory Information Management Systems (LIMS) developed in our laboratory. These LIMSs generate bar codes and text files that communicate directly with our robots and parallel every step of the Gateway recombinational cloning system from primer design through cloning, and ultimately sequence verification of full-length clones, thus ensuring a seamless flow of information between robots and LIMSs throughout (see figure). You can find more details about these systems here.
Collaborators and Projects
Glycoenzyme Repository: Kelley Moremen, Ph.D. (University of Georgia) and Don Jarvis, Ph.D. (University of Wyoming). PLASMIDS
Membrane Proteins in Infectious Diseases (MPID): Petra Fromme, Ph.D. (Arizona State University). PLASMIDS
PSI:Biology Plasmid Sequence Analysis: 4 High Throughput Centers, 8 Membrane Centers and 12 Biological Partners (view all Center names and affiliations here). PLASMIDS
Viral gene cloning: Ji Qiu, Ph.D. (CPD, Arizona State University)
Tuberculosis gene cloning: Mitch Magee, Ph.D.(CPD, Arizona State University) and Xiaobo Xu, Ph.D. (CPD, Arizona State University)
Other cloning projects: Shwetal Mehta, Ph.D. (Barrow Neurological Institute), Fridtjof lund-johansen, M.D., Ph.D. (Oslo University Hospital), Karen Anderson Laboratory (CPD, Arizona State University) and Marco Mangone's Laboratory (CPD, Arizona State University)
Publications
Seiler CY, Park JG, Sharma A, Hunter P, Surapaneni P, Sedillo C, Field J, Algar R, Price A, Steel J, Throop A, Fiacco M, Labaer J.(2013) DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2013 Nov 12. [Epub ahead of print] Abstract
Festa, F.; Steel, J.; Bian, X.; LaBaer, J., High-throughput cloning and expression library creation for functional proteomics. Proteomics 2013.Mar 4. doi: 10.1002/pmic.201200456. Abstract
Cormier CY, Park JG, Fiacco M, Steel J, Hunter P, Kramer J, Singla R, Labaer J. (2011) PSI:Biology-materials repository: a biologist's resource for protein expression plasmids. J Struct Funct Genomics. Mar 1 [Epub ahead of print]. July 1: 12 (2):55-62. Abstract
Cormier, CY; Mohr, SE; Zuo, D; Hu, Y; Rolfs, A; Kramer, J; Taycher, E; Kelley, F; Fiacco, M; Turnbull, G; LaBaer, J. (2010) Protein Structure Initiative Material Repository: an open shared public resource of structural genomics plasmids for the biological community. Nucleic Acids Research Jan;38(Database issue):D743-9. PMID: 19906724
Montor WR, Huang J, Hu Y, Hainsworth E, Lynch S, Kronish JW, Ordonez CL, Logvinenko T, Lory S, LaBaer J. (2009) Genome-wide study of Pseudomonas aeruginosa outer membrane protein immunogenicity using self-assembling protein microarrays. Infection and Immunity Nov;77(11):4877-86. Epub 2009 Sep 8.
Rolfs A, Hu Y, Ebert L, Hoffmann D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, Shen B, Baqui MM, Pearlberg J, Taycher E, DeLoughery C, Hoerlein A, Korn B, LaBaer J. (2008) A biomedically enriched collection of 7000 human ORF clones. PLoS ONE. 3(1):e1528. PMID: 18231609
Rolfs A, Montor WR, Yoon SS, Hu Y, Bhullar B, Kelley F, McCarron S, Jepson DA, Shen B, Taycher E, Mohr SE, Zuo D, Williamson J, Mekalanos J, Labaer J. (2008) Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. Proc Natl Acad Sci U S A. 105(11):4364-9. Epub 2008 Mar 12. PMID: 18337508
Taycher E, Rolfs A, Hu Y, Zuo D,Mohr SE, Williamson J, LaBaer J. A novel approach to sequence validating protein expression clones with automated decision making. BMC Bioinformatics. 2007 Jun 13;8:198. PMID: 17567908
Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, KelleyF, McCarron S, Jepson D, Richardson A, Raphael J, Moreira D, Taycher E, Zuo D, Mohr S, Kane MF, Williamson J, Simpson A, Bulyk ML, Harlow E, Marsischky G, Kolodner RD, LaBaer J. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007 Apr;17(4):536-43. Epub 2007 Feb 23. PMID: 17322287
Murthy T, Rolfs A, Hu Y, Shi Z, Raphael J, Moreira D, Kelley F, McCarron S, Jepson D, Taycher E, Zuo D, Mohr S E, Fernandez M, Brizuela L, LaBaer J. A full-genomic sequence-verified protein-coding gene collection for Francisella tularensis. PLoS ONE 2007;2:e577 PMID: 17593976
Park J, Hu Y, Murthy TV, Vannberg F, Shen B, Rolfs A, Hutti JE, Cantley LC, Labaer J, Harlow E, Brizuela L. Building a human kinase gene repository: bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci U S A. 2005 Jun 7;102(23):8114-9. Epub 2005 May 31. PMID: 15928075
Marsischky G, LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 2004 Oct;14(10B):2020-8. PMID: 15489321
Aguiar JC, LaBaer J, Blair PL, Shamailova VY, Koundinya M, Russell JA, Huang F, Mar W, Anthony RM, Witney A, Caruana SR, Brizuela L, Sacci JB Jr, Hoffman SL, Carucci DJ. High-throughput generation of P. falciparum functional molecules by recombinational cloning. Genome Res. 2004 Oct;14(10B):2076-82. PMID: 15489329
Labaer J, Qiu Q, Anumanthan A, Mar W, Zuo D, Murthy TV, Taycher H, Halleck A, Hainsworth E, Lory S, Brizuela L. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 2004 Oct;14(10B):2190-200. PMID: 15489342
Brizuela L, Richardson A, Marsischky G, Labaer J. The FLEXGene repository: exploiting the fruits of the genome projects by creating a needed resource to face the challenges of the post-genomic era. Arch Med Res. 2002 Jul-Aug;33(4):318-24. PMID: 12234520

